ArainGang, has posted a pretty interesting map of various ancestry components in the subcontinent by population. It’s pretty good, especially for the south and west of the subcontinent. But, there is something weird going on in the northeast: a lot of these populations have “Ancestral Indian” (Andamanese) ancestry but hardly anything else East Asian. This seems wrong. In fact, the Khasi are on a cline to Bengalis. I ran a few analyses on samples with the Andamanese and I just don’t see that Global 25 is doing this right.
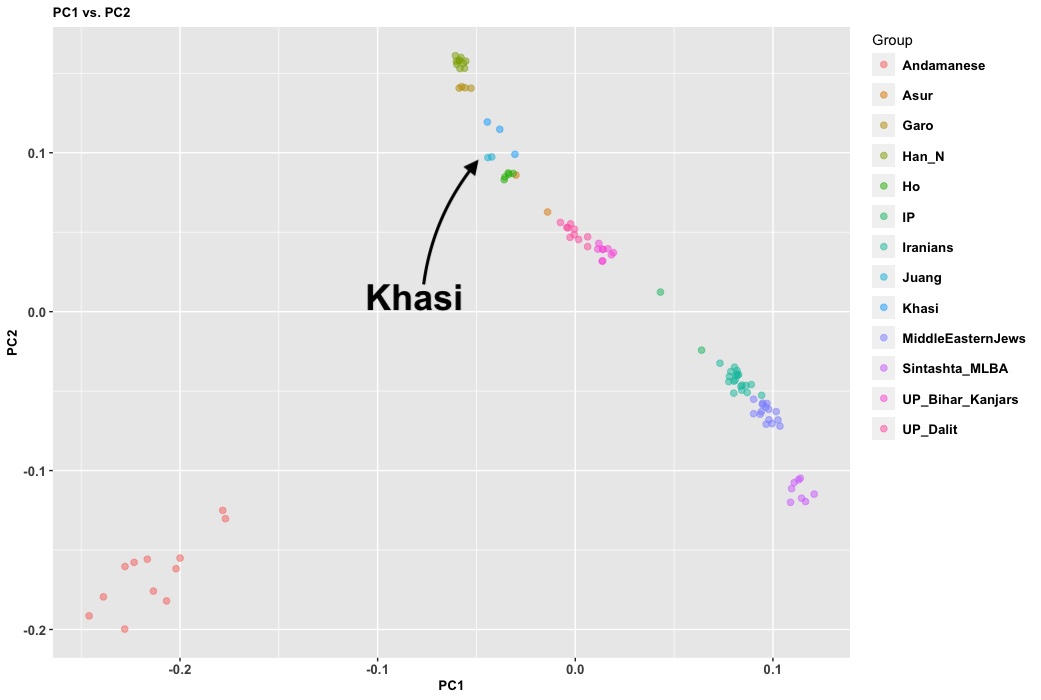
In the Global 25 model above the Khasi are 33% Ancient Indian, proxy for AASI, who are most closely related to the Andamanese. But you see in the analysis here the Khasi are along the India cline, but very shifted to the Han Chinese.
I ran a three-population test with a bunch of populations. You can see here that though the Andamanese are in the data set, the Khasi are best thought of as a mix of Han Chinese with an on-elite North Indian population.
| pop a | pop b | f3 stat | error | Z-score | |
| Khasi | UP_Dalit | Han_N | -0.0012727 | 0.000328938 | -3.8691 |
| Khasi | UP_Bihar_Kanjars | Han_N | -0.0010221 | 0.000334709 | -3.0537 |
| Khasi | IP | Han_N | -0.00120191 | 0.000481175 | -2.49787 |
| Khasi | Sintashta_MLBA | Han_N | -0.00080455 | 0.000392122 | -2.05179 |
What does this mean? I don’t think it’s a big deal. If the population does not have East Asian ancestry to a great extent the plot by Araingang looks fine. But, obviously, Global 25 has some kinks that people need to consider. This is important because people often come to me with Global 25 as if it’s authoritative. It’s not. It’s just another way to reduce genetic variation in a human consumable fashion.

recent genetic paper showing very high IVCp for Toda Tribes in South India,More than Kodavas and Panta Kapu
Toda
Indus_P: 0.707
AASI: 0.237
Steppe: 0.056
Gujjar
Indus_P: 0.714
AASI: 0.091
Steppe: 0.196
@ABCD, look at the p-values of the qpADM models especially for gujjars , Kodavas and panta_kapu. They are disgustingly low and well below the p-value threshold of 0.05. These are failed models, why cite estimates of these failed models as if they are accurate, authoritative estimates ?
30-35% of Punjab is dalits who cluster like dalits in other places. There are many more ethnic groups in many more of these places than what this map implies. Banias are a tiny proportion of Gujarat. Jats are a minority of Punjab and Haryana.
I also don’t understand the steppe discordance in this map vs. Narsimha et al 2018. There the steppe percentages were more tightly distributed.
Are you talking about the Punjab Jatt in the Narasimhan set? I have heard on anthrogenica that some of them were mislabeled and didn’t fit in the Punjab Jatt cluster. In global25, after using shahr sources, gonur ba1, paniya and sintashta, Punjab Jatts get high 20s, almost 30% on average.
No I’m talking about other groups. Many groups in Narsimha were like 15-20% steppe but are coming up a lot lower on here, like 0-5%. But groups like Khatris and Lohana were like 25-30% and coming up accurately here. So I don’t get that discordance.
How did S Indian Brahmins magically go from like 20% steppe in Narsimha to like 5% here.
I am not talking about the theories of Jats on anthrogenica. That’s too much of a quagmire to wade into.
30% is too much for the Khatri average. Model them as Shahr sources + Gonur + Sintashta + Paniya. This is a very bare bones model without any simulated inputs.
(without gonur)
Target: Khatri_o
Distance: 2.0226% / 0.02022589
55.8 IRN_Shahr_I_Sokhta_BA2
24.6 RUS_Sintashta_MLBA
16.6 Paniya
3.0 IRN_Shahr_I_Sokhta_BA1
Target: Khatri
Distance: 1.6512% / 0.01651173
33.6 IRN_Shahr_I_Sokhta_BA2
28.8 IRN_Shahr_I_Sokhta_BA1
23.8 RUS_Sintashta_MLBA
13.8 Paniya
(with gonur)
Target: Khatri_o
Distance: 2.0196% / 0.02019554
56.0 IRN_Shahr_I_Sokhta_BA2
24.2 RUS_Sintashta_MLBA
16.6 Paniya
3.2 TKM_Gonur1_BA
Target: Khatri
Distance: 1.6156% / 0.01615614
39.2 IRN_Shahr_I_Sokhta_BA2
22.6 RUS_Sintashta_MLBA
17.6 TKM_Gonur1_BA
12.2 Paniya
8.4 IRN_Shahr_I_Sokhta_BA1
I do not have Lohana coordinates so I can’t make concrete comments on that estimation but if it is anything like this, then it could be an overestimation as well.
>South Indian Brahmins
Target: Brahmin_Tamil_Nadu
Distance: 2.2098% / 0.02209775
57.6 IRN_Shahr_I_Sokhta_BA2
26.6 Paniya
14.4 RUS_Sintashta_MLBA
1.4 IRN_Shahr_I_Sokhta_BA1
0.0 TKM_Gonur1_BA
Yeah they aren’t 5% in this model, something like 15%.
>too much quagmire
You can talk to paindu directly if he shows up here.
Yes so why is this graphic underestimating groups with 5-15% steppe and not groups with 20-30% steppe. I have a suspicion that the guy is playing games to select things that maximize that difference. It’s just based on priors. Like it looks off
I think there are more issues than just the E Asian samples. I think basically only the NW seems to be consistent with other models.
People do that a lot recently. They re-name components differently from their original global25 sheet names. Do not mention which ones are simulated, and in some cases, even in vahaduo screenshots they have the gall to hide the fits. No point in using a tool that is more accurate than gedmatch calculators if they are just going to drag it down to that accuracy level.
@warlock, not really with respect to punjabi dalits clustering with other dalits of other region. With respect to punjabi dalits, it depends upon the caste. Punjabis Cham*rs seem at least as west eurasian shifted as south indian brahmins based on Harappaworld, G25 etc. It’s the valmikis who are more south-shifted.
Even after removing jats, brahmins, khatris from punjab, other castes like sainis, gujjaes, tarkhans are also quite west eurasian shifted(By west eurasian , i mean of combo of Iran_N + Sreppe).
Yes but the way this looks, that huge cluster that is even S Indian Brahmin like let’s say just doesn’t show up. The guy takes the most West Eurasian shifted in the Indus Plain and tends to compare it to the least West Eurasian shifted elsewhere, except for some Brahmins. There needs to be all groups with weighted averages or just show all groups.
In general, many groups show some steppe in Narsimha and show pretty much none here. But the steppe in NW group stays consistent. Also this Middle Eastern isn’t really in Narsimha. This seems odd.
He has an agenda of a a “separate Indus nation.” That’s fine if he was accurate with showing everything. But he isn’t he spins everything a lot, including not including Gujarat as a successor of Indus. He likes Harayana because of higher steppe and loves to include them. Hence, why he posts Chopra javelin stuff more than a couple times. It fits his narrative of some NW Indus Master Race Vs. Rest, especially shitty Gangus. The irony is that he is half gangetic himself. Basically, he lets his motivated reasoning show. And that shows with how he presents data. Look at home use open defecation numbers from 2011 in 2021 to shame India. It’s the same thing her. Subtle modifications to fit a narrative. There is some gaslighting here. It’s still a decent map. But there are some obvious choices made here.
Gujjars of Rajasthan show close to SW Asian on Harrappa like Mers, but look at the numbers here. Similarly, the numbers are way off for, SI Brahmins, Gangetic Brahmins, Rawats, Chhettris, HP Rajput, kshatriya etc. Are we being told that Gangetic Brahmin carry less Steppe than Lohana/Khatri/ or non-Jat/Ror grp of NW. Absolutely joke. I do agree with the point
@TD
Chamars are diverse but all the samples, except for 1, do not cluster as far north as you claim. They cluster with general S Indian populations well. If that 30-35% of Punjab clusters like that, then this map is extremely misleading. It neglects giant chunks of society in all of these geographies. Looking at this guy’s past paradigm, it seems like a partial Biradri ethnosupremacist stunt. The plausible deniability is brilliant. This guy makes for a Class A propagandist. The ISI needs to hire him immediately to rewrite the Pak Studies curriculum. I am sure he is already in touch.
Target: Chamar:A260
Distance: 3.7861% / 0.03786123
63.0 Paniya
34.4 IRN_Shahr_I_Sokhta_BA2
2.6 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Chamar:A259
Distance: 3.5712% / 0.03571233
56.8 Paniya
38.8 IRN_Shahr_I_Sokhta_BA2
4.4 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Chamar:A261
Distance: 2.7761% / 0.02776055
58.6 Paniya
39.2 IRN_Shahr_I_Sokhta_BA2
2.2 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Chamar:evo_40
Distance: 2.4368% / 0.02436787
45.8 Paniya
39.0 IRN_Shahr_I_Sokhta_BA2
15.2 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Chamar:evo_41
Distance: 3.0211% / 0.03021093
63.2 Paniya
32.8 IRN_Shahr_I_Sokhta_BA2
4.0 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Chamar:evo_42
Distance: 3.4674% / 0.03467443
57.8 Paniya
40.6 IRN_Shahr_I_Sokhta_BA2
1.6 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
“Chamars are diverse but all the samples, except for 1, do not cluster as far north as you claim.
If that 30-35% of Punjab clusters like that, then this map is extremely misleading”
@warlock, I was talking about *PUNJABI chamars*, as far as I know, the ones whose data you posted are UP ones, again, I was talking about punjabi not central-east UP ones.
There are a few samples of Punjabi chamars on anthrogenica and they are more north shifted than the UP ones.
An Example is something like this , posting HarappaWorld Score ( I know HW is outdated and doesn’t tell estimates for source populations which gave rise to this population. but still an OK tool for comparison across populations and certain components like S-Indian do correlate with AASI)
Punjabi Chamar:
Population Percent
1 S-Indian 41.21
2 Baloch 35.76
3 Caucasian 10.58
4 NE-Euro 8.57
5 NE-Asian 1.27
6 Beringian 1.22
7 Mediterranean 1.17
8 San 0.23
For their G25 coordinates, I will ask anthrogenica guys.
Yeah. This stuff is so messy relying on alleged ancestries of individuals with people questioning validity regardless of what direction things go.
I concede you may be correct with the geographic change. But I don’t think we have a ckear answer unless there is a study with wider scale sampling with verified ancestry. This random submission stuff or contested samples in official literature is too messy. There is too much both motivated reasoning and potential mislabeling. And it will only get messier as groups mix more, a good thing I think.
The other problem is that of subcastes. Every caste has a crap ton of different subdivisions. Some are considered higher or lower. Some intermarry some don’t. It’s a mess
Also, out of curiosity. Are they verified UP ones? I am curious how people concluded that (could be valid but curious of source, so as to verify that it isn’t made up by some people who just want to confirm priors, all too common in this space)
“I concede you may be correct with the geographic change. But I don’t think we have a ckear answer unless there is a study with wider scale sampling with verified ancestry. This random submission stuff or contested samples in official literature is too messy. .”
Yeah, we need proper academic samples. But fwiw, I have an HarappaWorld sheet where there are 10 punjabi chamar samples. Their SI ranged from 40% to 47%. The average of the 10 samples was something like this –
Punjabi Chamar average:
1 S-Indian 42.72%
2 Baloch 35.26%
3 Caucasian 7.63%
4 NE Euro 7.91%
5 SE Asian 0.31%
6 Siberian 1.01%
7 NE Asian 0.79%
8 Mediterranean 1.11%
“The other problem is that of subcastes. Every caste has a crap ton of different subdivisions. Some are considered higher or lower. Some intermarry some don’t. It’s a mess”
Yeah, I agree, for some castes, subcastes can be another axis for variation. I full expect variation among the UP and bihar yadav subcastes.
“Also, out of curiosity. Are they verified UP ones? I am curious how people concluded that (could be valid but curious of source, so as to verify that it isn’t made up by some people who just want to confirm priors, all too common in this space)”
Afaik, they were from metspalu papers which had samples from UP. Also, from what I know( and I may be wrong) the samples present in https://genoplot.com/admix are the samples which were collected in academic papers. If you want to check for private samples, you may go at https://genoplot.com/g25. Here, there’s a sample named ‘User – Duffydemon Chamar (2020 CE Punjab)’. You can compare this with ‘Brahmin Tamil Nadu Average’ and ‘Chamar Average’ on a custom G25 calculator ( I used ‘South Asian Bronze Age(Scaled) by HekSindhi’).
You can see the results here at https://shared.genoplot.com/file/genofiles/dante911/table-181ee73e56a
(IndusPeriphery is Shahr-i-Sokhta BA3,
Excess East Eurasian is Chokhopnai 2700BP,
Steppe_MLBA)
You can see how that duffydemon guy is similar to TN Brahmin average and quite north-shifted compared to those chamar average.
Actually they are about as North shifted as N Indian Ksyhtrias with only the most South shifted of those samples being like Tam Brahms. Interesting.
Again, makes you think, are there different subpopulations etc.
not being a geneticist (?), all i can see is that the dravidian brahmin and the konkani brahmin are almost same. what ever happened to the nordic origins of these konkani brahmins with their green eyes??!!
Sexual selection and relative inbreeding is magical
That’s partly why, not just climate, why to the common Indian, Kashmiris look more West shifted on average than even the most West shifted Punjabi populations, who are on average definitely more West Shifted than the average Kashmiri, albeit by not much but certainly not a negligible amount.
The problem here is using simulated populations and distant ones like “ancient iranians” instead of more proximal ones like shahr. And the issue doesn’t seem to be limited to Khasi only.
not being a geneticist (?), all i can see is that the dravidian brahmin and the konkani brahmin are almost same. what ever happened to the nordic origins of these konkani brahmins with their green eyes??!!
made up.
The problem here is using simulated populations and distant ones like “ancient iranians” instead of more proximal ones like shahr. And the issue doesn’t seem to be limited to Khasi only.
yeah, every east asian enriched group in this list has more ‘ancient indian’ than it should, and i think that’s due to east asian => ancient Indian shift.
Looking at the Maratha, Lingayat and Vokkaliga proportions, I am surprised that Ancestral Indian is highest in Maratha followed by Lingayats and Vokkaligas. For Ancient Iranian it is the reverse. Is this difference mean anything or could be attibuted to sampling error?
@Bhumiputra could be that there was movement of people from Karnataka dried regions to Maharashtra, with Maharashtra region having population with more AASI ( may be some areas in Maharashtra were not explored).
@Razib
@Dathang
@TD
Ok here are some clear examples of some discordance with Reich Lab Data. For NW groups, it looks like it is concordant. For other groups, looks like possible steppe underestimate, looking at these pie charts.
NW groups (steppe here matches)
Lohana 28.1
Khatri 26.9
Other groups and related that look way lower on this chart but higher in Narsima for Steppe MLBA
Yadav: 22.6
Kurmi 16.4
Brahmin Karnataka: 19.6
Baniya 16.4
Maratha 11.1
Ligayat 10.9
Brahmin Catholic of Kerela 14.3, 13.7
Corgi 9.8
Kapu 6.4
What explains this discordance? If in fact, Reich lab numbers are accurate- the difference in steppe between NW and the rest is not as pronounced as this graphic communicates.
@Warlock, I am not a geneticist but I will try to explain the discordance. He has used G25 (especially simulated populations like AASI which he got by subtracting east asian from Onge and Ancestral Iranian which he got by subtracting AASI from IVCp) instead of qpAdm. I believe such choices simulated/ghost ancestral populations and use of G25 caused divergence here.
I have seen G25 estimates diverging from passing qpADM model estimates even when both used the same source populations.
Regarding Khatri and Lohana estimates, well, the thing is qpAdm models validity need to be judged by their p-values. Check out the p-values for Khatris and Lohanas in narasimhan’s paper (and in the new paper). They are extremely bad. This means that Khatris and Lohanas can NOT modelled using Onge, IVCp
and Steppe_MLBA. Something else is needed for them.
Model any of these groups with the following sources on your own on the vahaduo g25 website: Shahr BA1, Shahr BA2, Sintashta MLBA, Gonur BA1 Paniya. Add tyumen in a separate run. Do not use any simulateds, and see what the results are. I think his Pashtun ethic group and Kho results have too much steppe. They get 30% with the aforementioned model.
Target: Kho_Singanali
Distance: 3.6748% / 0.03674773
29.6 RUS_Sintashta_MLBA
27.2 TKM_Gonur1_BA
24.4 Paniya
17.8 IRN_Shahr_I_Sokhta_BA1
1.0 IRN_Shahr_I_Sokhta_BA2
Target: Pashtun_North_Afghanistan
Distance: 2.6533% / 0.02653285
51.6 TKM_Gonur1_BA
29.4 RUS_Sintashta_MLBA
19.0 Paniya
0.0 IRN_Shahr_I_Sokhta_BA1
0.0 IRN_Shahr_I_Sokhta_BA2
Target: Pashtun_Kurram
Distance: 1.6352% / 0.01635159
33.0 TKM_Gonur1_BA
26.6 IRN_Shahr_I_Sokhta_BA2
24.4 RUS_Sintashta_MLBA
16.0 Paniya
0.0 IRN_Shahr_I_Sokhta_BA1
The Punjab Jatt accuracy however cannot be checked/corrected for without removing the outliers that don’t cluster and are possibly mislabeled. I do not know which these samples are, so for that you’ll have to talk to Paindu and some other anthrogenica people.
Yes. When you do, the numbers look higher East and South of the NW for groups than what he has portrayed on these pie charts. It’s a subtle thing but it makes the difference look bigger than what it is. The E Asian thing looked less deliberate. But knowing what we know about his biases, I suspect he is messing around to portray a wider gap than there is.
Razib says average NW/Pak steppe is 25%. Average N India is 15%. Average S India is 5%. This guy makes it look like 30% in NW, 5% in N, and 2% in S. Something is off
Ok I did some runs for the populations. The map looks discordant. Assume BA2 is like 30% AASI
Target: Warlock (can substitute for Bania)
Distance: 3.1757% / 0.03175669
60.2 IRN_Shahr_I_Sokhta_BA2
27.0 Paniya
12.8 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Kshatriya:195
Distance: 2.6592% / 0.02659219
47.0 IRN_Shahr_I_Sokhta_BA2
32.8 Paniya
13.6 RUS_Sintashta_MLBA
6.6 IRN_Shahr_I_Sokhta_BA1
0.0 TKM_Gonur1_BA
Target: Brahmin_Tamil_Nadu:SB001
Distance: 2.3857% / 0.02385730
58.0 IRN_Shahr_I_Sokhta_BA2
26.8 Paniya
13.8 RUS_Sintashta_MLBA
1.2 IRN_Shahr_I_Sokhta_BA1
0.2 TKM_Gonur1_BA
Target: Gujar_India:GJ16
Distance: 2.5604% / 0.02560442
67.2 IRN_Shahr_I_Sokhta_BA2
19.4 RUS_Sintashta_MLBA
8.2 Paniya
5.2 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Gujar_Pakistan:G-168
Distance: 2.1967% / 0.02196666
59.2 IRN_Shahr_I_Sokhta_BA2
22.8 RUS_Sintashta_MLBA
13.6 IRN_Shahr_I_Sokhta_BA1
4.4 Paniya
0.0 TKM_Gonur1_BA
Target: Brahmin_Uttar_Pradesh:BR052
Distance: 4.3797% / 0.04379722
49.6 IRN_Shahr_I_Sokhta_BA2
26.0 Paniya
24.4 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Khatri_o:K-123
Distance: 2.6221% / 0.02622119
58.4 IRN_Shahr_I_Sokhta_BA2
25.4 RUS_Sintashta_MLBA
16.2 Paniya
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Kamboj:KJ_03
Distance: 2.9210% / 0.02920968
41.4 IRN_Shahr_I_Sokhta_BA1
26.8 RUS_Sintashta_MLBA
16.4 IRN_Shahr_I_Sokhta_BA2
15.4 Paniya
0.0 TKM_Gonur1_BA
Target: Punjabi_Jatt:PJ004
Distance: 2.7901% / 0.02790100
57.2 IRN_Shahr_I_Sokhta_BA2
27.2 RUS_Sintashta_MLBA
10.4 TKM_Gonur1_BA
5.2 Paniya
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Ror:Ror_10
Distance: 2.3529% / 0.02352876
41.2 IRN_Shahr_I_Sokhta_BA2
38.8 RUS_Sintashta_MLBA
10.2 IRN_Shahr_I_Sokhta_BA1
9.8 Paniya
0.0 TKM_Gonur1_BA
Target: Sri_Lankan:GRC10041304
Distance: 2.7638% / 0.02763842
57.2 IRN_Shahr_I_Sokhta_BA2
39.8 Paniya
3.0 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Konkani_Christian:12701
Distance: 2.7866% / 0.02786615
52.6 Paniya
41.4 IRN_Shahr_I_Sokhta_BA2
6.0 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Velamas:VELZ260
Distance: 2.1549% / 0.02154910
68.8 IRN_Shahr_I_Sokhta_BA2
26.2 Paniya
5.0 RUS_Sintashta_MLBA
0.0 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Target: Yadava:S_Yadava-1
Distance: 2.8847% / 0.02884707
53.4 Paniya
40.8 IRN_Shahr_I_Sokhta_BA2
5.0 RUS_Sintashta_MLBA
0.8 TKM_Gonur1_BA
0.0 IRN_Shahr_I_Sokhta_BA1
Regarding discordance between admixture from my chart and admixtures published in certain papers:
A lot of papers use odd reference populations for their admixture calculations. Specifically, using Iran-N or IVC to approximate the Iranian HG component in Indians. This leads to significant artificial inflation of Steppe across all populations.
Sampling is also an issue. Where the samples come from (there are academic Ahir/Yadav samples floating around, but they are from Haryana, while Ahir/Yadav I use is UP). And the sample size (some averages on my chart are comprised of 1-5 individuals, where an academic may have dozens).
On Twitter and my Medium page I posted what reference populations I used, anyone can go run them on Genoplot to check if any shenanigans were going on with the Steppe scores. Andamese+Iran Hotu+ Hajji Firuz C+ Sintashta.
Lastly, the populations for this chart were carefully selected to be as representative as possible, and generally are members of the largest caste/tribe for their respective region.
Please post the exact percentages in numbers for each component, the graph is good but we need numbers for accuracy
If sampling is an issue and you have 1-5,often only self reported samples, then don’t you question the integrity of this? Do you find it convenient that you happen to use a fit that lowers steppe appreciably across the board except for the NW groups? What pan inflation? Gujjars, Aroras, Lohanas, Jats, and Rors look nearly constant to other models. But everyone else, including Brahmins drops? Something seems off.
I’ll wait for Razib to weigh in too.
As far as integrity goes, I’m mostly concerned with whether this is better than anything else available (it is), rather than if its perfect (it is not). Fwiw I don’t think more samples would appreciably change the scores of most populations I tabulated.
NW groups also have Steppe come down in my model. For example, the Lohana scored 19.5% Steppe on my chart, compared to 29.5% if I run the average with Iran-N or IVC rather than Iran Hotu.
I’m also not too worried about this, as I’m confident this is the right way to model South Asians, and that earlier papers were using poor reference populations leading to ridiculously elevated Steppe estimates. You can see this even when trying to model IVC with Iran-N rather than Iran-Hotu, which gives IVC nearly 5% Steppe when they should have 0%.
I do not agree with your proposition. Your model distances are far worse than what Dathang proposed. Just compare.
Target: Warlock
Distance: 11.1394% / 0.11139412
50.0 IRN_HotuIIIb_Meso
43.6 IND_Great_Andamanese_100BP
6.4 RUS_Sintashta_MLBA
0.0 IRN_Hajji_Firuz_C
Target: Ror:Ror_10
Distance: 6.2281% / 0.06228090
41.8 IRN_HotuIIIb_Meso
34.8 RUS_Sintashta_MLBA
23.4 IND_Great_Andamanese_100BP
0.0 IRN_Hajji_Firuz_C
Target: Punjabi_Jatt:PJ003
Distance: 6.7381% / 0.06738080
49.0 IRN_HotuIIIb_Meso
24.8 RUS_Sintashta_MLBA
24.8 IND_Great_Andamanese_100BP
1.4 IRN_Hajji_Firuz_C
Target: Khatri_o:K-123
Distance: 9.4160% / 0.09415958
45.4 IRN_HotuIIIb_Meso
34.2 IND_Great_Andamanese_100BP
20.4 RUS_Sintashta_MLBA
0.0 IRN_Hajji_Firuz_C
Target: Brahmin_Tamil_Nadu:SB003
Distance: 9.9662% / 0.09966231
49.2 IRN_HotuIIIb_Meso
42.8 IND_Great_Andamanese_100BP
8.0 RUS_Sintashta_MLBA
0.0 IRN_Hajji_Firuz_C
Target: Brahmin_Uttar_Pradesh:BR008
Distance: 11.1704% / 0.11170390
47.8 IRN_HotuIIIb_Meso
38.0 IND_Great_Andamanese_100BP
14.2 RUS_Sintashta_MLBA
0.0 IRN_Hajji_Firuz_C
Target: Yadava:S_Yadava-1
Distance: 12.6453% / 0.12645299
59.6 IND_Great_Andamanese_100BP
40.4 IRN_HotuIIIb_Meso
0.0 IRN_Hajji_Firuz_C
0.0 RUS_Sintashta_MLBA
Target: Chamar:A261
Distance: 13.7810% / 0.13781039
62.4 IND_Great_Andamanese_100BP
37.6 IRN_HotuIIIb_Meso
0.0 IRN_Hajji_Firuz_C
0.0 RUS_Sintashta_MLBA
Target: Konkani_Christian:12701
Distance: 13.1408% / 0.13140810
58.6 IND_Great_Andamanese_100BP
41.4 IRN_HotuIIIb_Meso
0.0 IRN_Hajji_Firuz_C
0.0 RUS_Sintashta_MLBA
Target: Kshatriya:198
Distance: 10.1590% / 0.10158986
45.4 IRN_HotuIIIb_Meso
41.2 IND_Great_Andamanese_100BP
13.4 RUS_Sintashta_MLBA
0.0 IRN_Hajji_Firuz_C
Yup that’s another thing some people do- they use an obscure combo which naturally produces bad fits. Not as bad as the screenshots directly from vahaduo where people don’t even post the fits, buy people need to be cognescent that a distance way bigger than 3 isn’t telling much, and beyond that point the bigger it is, the less meaningful it is.
@araingang Can you post the scaled Lohana coordinates here?
I think diaspora commentators are grossly overestimating how much race matters in the subcontinent for political and national identities. The groups in India that are the most hostile to Pakistan are precisely the NW groups (Punjab and Haryana) and Brahmins across the board. Actual non-Steppe heavy regions and groups mostly dont care too much, and actually seek to maintain their own distance with Delhi.
Similarly, groups like Muhajirs in Pakistan, which have more non-Steppe ancestry than the locals, practically led the Pakistan movement and have guided its identity and policies.
The subcontinent is absolutely saturated with religious symbolism and culture. Even today, most Bangladeshis and Pakistanis will say they are Muslim before anything else. India has open borders with Hindu majority Nepal. Indian Dalits are willing to vote with the BJP, which is upper caste heavy because they share the aversion to Muslims.
It is unclear that genetics provides any guidance to actual politics in the subcontinent. That diaspora commentators care so much about it reflects their own experience growing up in societies with a legacy of explicit racism and a continuing tradition of racially ascribed groupings.
‘I think diaspora commentators are grossly overestimating how much race matters in the subcontinent for political and national identities.’
They know. They just dont care. Just like Indians as such dont care about these matters.
The last thing we need in India is a genuine racial divide. I am already coming across second generation Indians identifying as ‘Dravidian nationalists’. Remember these folks can get OCI cards and are dollar rich Americans.
They are seeking a catharsis from their racist experience in the US via a vivisection of a faraway country with better ideals.
Read some trad accounts and your jaw will drop. They are worse supremacists than some of the most Jatt Sikh supremacist Khalistanis.
There is a theme in general of feudal land owning castes who failed to adapt to modern economies, having a chip on their shoulder about the system, especially when they are dependent on very useful but state subsidized jobs (eg. Farming and army).
Some groups like Patels and Reddy have imitated mercentile groups well. Others like Thakur have not. Some of the most powerful are doing so because they have a ton of starting capital. But they seethe from their rented out palaces, as the subaltern rises.
This is a minority. But it exists. And it reigns in excess concessions by mainstream Hindutuva. But it is an ugly demon. A lesser version of what TTP is like to Pak. Sadly, these feudal types are who run Pak like PPP.
I will praise Imran. Because he at least paid lip service to changing it. And frankly more, with his pivot to China and praise of the notion of eventually creating a welfare state. This is a big reason for his ouster.
But like I know this racial stuff is mostly just a small layer or inconsequential to the Indian psyche, you already know all of this.
But remember. Every time someone brings up “fair skin” and “sharp features,” they are indirectly referring to this stuff. Studying it to understand Indian bias is another reason I’m into it. Because it explains so much of real and casual prejudice, ofcourse incompletely, but at least it gives big clues.
That’s a sad view of history. I don’t think my direct ancestors likely did anything super substantial. They were probably small time merchants, at best. But caring so much about that is weird.
History is history. I think this lineage obsession is the problem. Living in east glory too much because India is relatively so poor today. And S India has some great Kingdoms so who cares. And it is doing the best out of all regions today.
We all speak languages, follow religions, and have cultures that are foreign to some level. Who cares. Find out what idealogy best suits you and do that.
India needs some of the rational individualism of the West. Yes, the confederacy of lineage and caste can be protective. But in the modern era with a globalizing world focused on tech and markets, it is a net hindrance.
“The Left is stuck in the past:” is a literally true allegory.
“ I am already coming across second generation Indians identifying as ‘Dravidian nationalists’.
I have come across more Indians in india who identify as “ Dravidian nationalist”. If u haven’t noticed, they have a political party. Which like already rules a state.
Most of the this racial thing in India is mostly driven by inferiority complex of left – Dravidian folks who have nothing original to claim of. They see most of what’s seen as Indian culture is mostly North Indian culture. Plus the religion they follow also happens to be North Indian. Plus unfortunately for them, even the indus- harrapa thing ( which the Dravidian wanna claim ) also falls in these wretched northern plains.
So they cling on to all this racial theory, because that’s they only thing they have to separate themselves. And to show that they are the OG while the northies are interlopers.
It’s a coping mechanism, so let them cope.
Common Saurav, do you really think South Indians havent done anything substantial ? Above all, I actually think they are one of the more sensible groups of humans on this planet. There has to be a reason why South India has not seen a war for nearly 300 years.
@thewarlock, I dont consider trads a threat. Feudals of the type you see in Pakistan dont havent existed in India for a few decades now. Land reform in the North, though not as extensive as in Kerala and Bengal (and J&K surprisingly) did happen.
S-Indians have done a lot of substantial things. Just not in the areas they want to claim. On the contrary, i feel N-Indians have been sitting on their bums for last 300 odd years, whereas S-Indians have zipped right passed through.
And i agree they are the perhaps the most sensible of all Indians. Which actually ties into the reason why the havent seen wars in last 300 years. The very same reason Bengal didn’t see wars after the battle of Plassey.
If you collaborate with the colonists, then there is nothing to fight for, isn’t it? Then the colonists also develops your region and throws few bones for you the chew on, here and there. Wars are fought against people who resist, No?
A pretty similar debate was had between Dathang and a former poster. I will try to find it and link it
” anyone can go run them on Genoplot to check if any shenanigans were going on with the Steppe scores. Andamese+Iran Hotu+ Hajji Firuz C+ Sintashta.
that earlier papers were using poor reference populations leading to ridiculously elevated Steppe estimates. ”
@ArainGang, not sure if you’ll see this but unlike G25 , qpAdm doesn’t allow for creation of ghost/simulated populations so researchers got to work with whatever samples they’ve got. They also can’t model modern groups with source pops like Sintastha and Iran_Hotu together because that would be completely anachronistic. All in all, G25 is not an academic tool so canceling previous research papers models (which were constrained by lack of appropriate samples) based on estimates obtained from simulated populations is a bit funny.
I understand that for scientists like Razib, genetic make-up holds clues to the population structure and history of the subcontinent. And for some weirdos of a chauvinistic bent, “steppe %” might make them feel good about belonging to a certain group.
I’m all for studying and gathering knowledge for knowledge’s sake, but outside these narrow interests is there anything of practical use that this sort of information will help unlock for scientists or medical practitioners? Like disease propensity, improving nutrition, etc.
As someone in the field, not really. This more global ancestry stuff means not a whole lot usually for patient care. Like for example, indigenous American Gene tend to make one susceptible to gallstones. Most modern day S Americans at least have some. Betting one way or the other is dumb. Why go off the surrogate of the overall ancestry. Just screen for the trait directly.
The GWAS stuff for individual traits related to disease and then more meta stuff on how they interact matters. Personal genomics will be huge in the future. Already for cancer, doctors prescribe different therapy to target your individual mutations. This will happen eventually with other major diseases (heart diseases diabetes, etc.)
But this stuff is fun because history, politics, etc is fun. Knowing more for the sake of knowing is good but it also helps inform the world today, in the right context. And this stuff definitely has relevance that way.
Fascinating, thanks for the info. Personal genomics and GWAS sounds really interesting as I’m a bit of a data and stats nerd.
“..it also helps inform the world today, in the right context”
True, but in the wrong hands this stuff can be toxic. The world needs less identity politics not more of it.
Yes, second Siddarth on this. Personalized medicine via genomics makes a lot of sense. Hopefully will also help children who have rare illnesses.
It already has. CF has been transformed with Ivacaftor. Sickle Cell and thalassemia are next. Basically point mutation diseases first ones to target
I’ve been involved with that type of research on the basic level. Got into computational side more this year with some Cancer Genome Atlas stuff. Slowly learning more. It’s cool stuff.
The labeling on this chart is either mischievous, malicious or downright ignorant.
– “Ancestral Indians” peak in Orissa/Jharkhand which is also the locus of Austro-Asiatic languages.
– IVC which is attested both archaeologically and genetically does not get a label.
– No archaeological attestation for “Ancestral Indians” or “Steppes Aryans”
– Which clade is the proxy for Dravidian languages – Ancestral Indian or Ancient Iranians? Both the Southworth & Krishnamurti models are at loggerheads in this representation.
Some of these inconsistencies would get resolved if the IVC Cline was showed as an ancestry source. But knowing the proclivities of the author, it is clear why this couldn’t be!
https://www.thehindu.com/news/cities/Madurai/theory-of-race-was-created-to-divide-people-claims-governor/article65636436.ece?homepage=true
Theory of race was created to divide people, claims Governor
‘He said the British learnt about the waterproofing of ships from the Cholas, and India was a great maritime power. Similarly, they gained other technical know-how and did not stop at that, but also killed the indigenous industry. They also killed the indigenous education system and introduced their rote-learning method so that people could serve as servants of their company, the Governor said.
When the “fire of freedom” started in the country, they created literature about Aryans and Dravidians being races, whereas this was just a geographical distinction, he said. He urged the people not to get carried away by such literature, and instead go to the archives and learn about these issues themselves.’
“Ancestral” Indian label is a bit confusing.
Why not use “Ancient Indian” like author did for “Ancient Iranian” since most Indic ppl have a lot of ancient Iranian esp in Western India and Indus region (modern day Pakistan).
His model is subpar. Look at the distances between his and Dathang’s proposed population modeling. His idealogy is showing through. He wants to max out certain components and show certain groups strategically to better justify partition racially. He is playing the usual “Indus Gang” games. And his labeling of components differently is intention and part of the jig. The guy is good at lawyering. His decisions are quite strategic.
are there any northeast indian G25 coordinates available publicly? for khasis, nagas, mizos, meiteis, nyishis, garos, etc. I’ve only seen jamatias/tripuris, manipuri brahmins on eurogenes G25 sheet but not the others. I’ve tried finding them online and I couldnt find any. It would be nice if their coordinates could be shared, especially khasis, or some assamese groups like chutias, rajbanshis, kalitas, etc.